Bvalcalc Documentation
Welcome to the documentation for Bvalcalc, a CLI tool for easily calculating B-values and B-maps.
Background selection (BGS) is the process by which diversity is reduced at neutral sites linked to conserved regions such as exons that experience direct purifying selection, and across the genome. Bvalcalc allows users to easily calculate the relative reduction of diversity due to BGS (B) using several different modes. Calculating B-values is important for understanding the linked effects of selection, can be used to establish an evolutionary null model for inference, and can aid in selecting the most neutrally evolving sites, see Demographic Inference with B-map.
To learn more about background selection, see About BGS and B-values. To access pre-built B-maps for select species, see Download B-maps. To calculate B-values for yourself, install Bvalcalc and explore the docs, we recommend getting started with the Quickstart Tutorial.
Bvalcalc [--generate_params [SPECIES]|--site|--gene|--region|--genome]
--generate_params [SPECIES]
Save population parameters from a species template (selfing, dromel_cds, dromel_utr, dromel_phastcons, homsap_cds, homsap_phastcons)
--site, -s Calculate B values for a single site from a selected element
--gene, -g Calculate B values for a region adjacent to a single selected element
--region, -r Calculate B values for a chromosomal region, considering genome-wide effects
--genome, -w Calculate B values genome-wide for all sites considering all elements
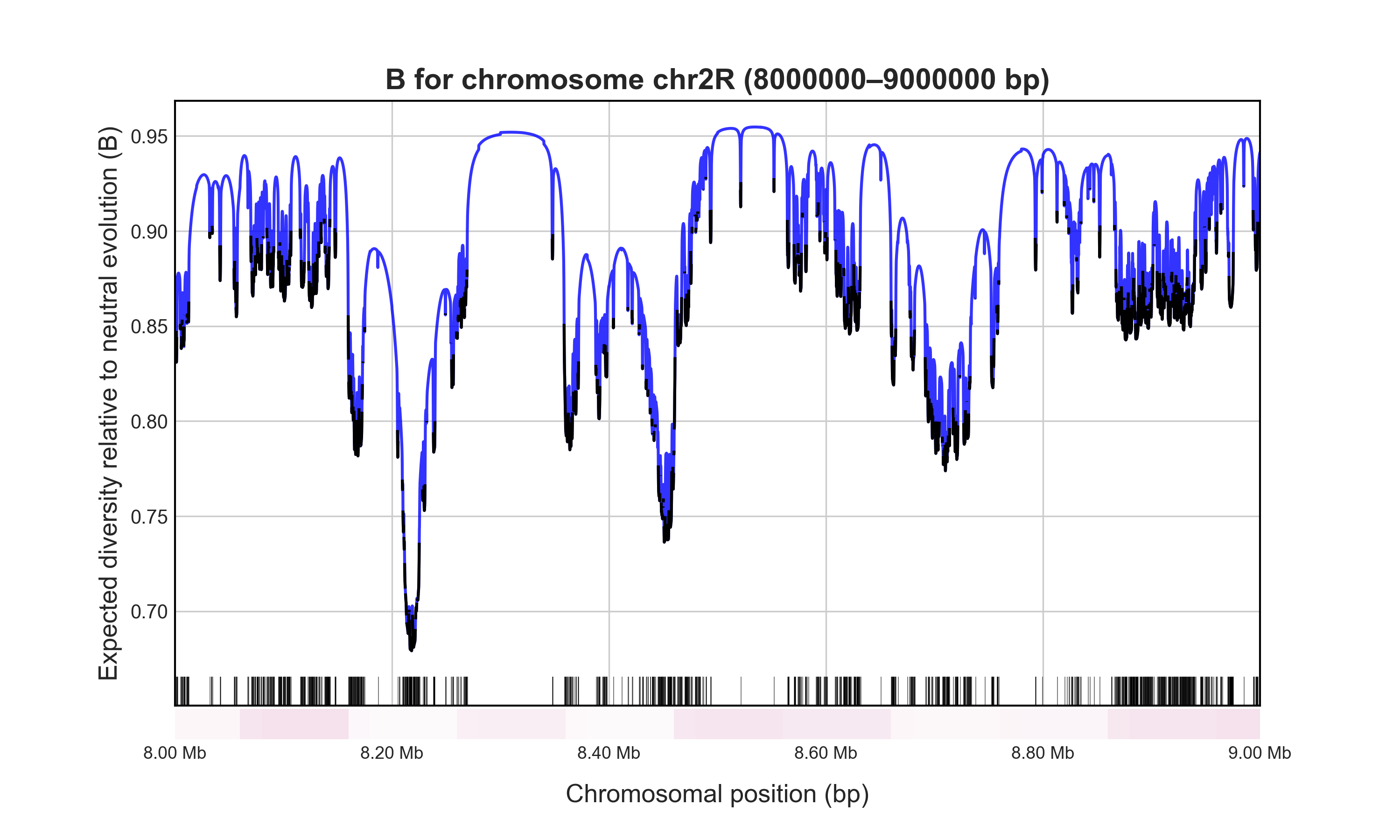
B-values at 1 Mb of sites under BGS from coding sequences in D. melanogaster, calculated with the Region module in 6.67 seconds
Introduction
Modules
Guides
API